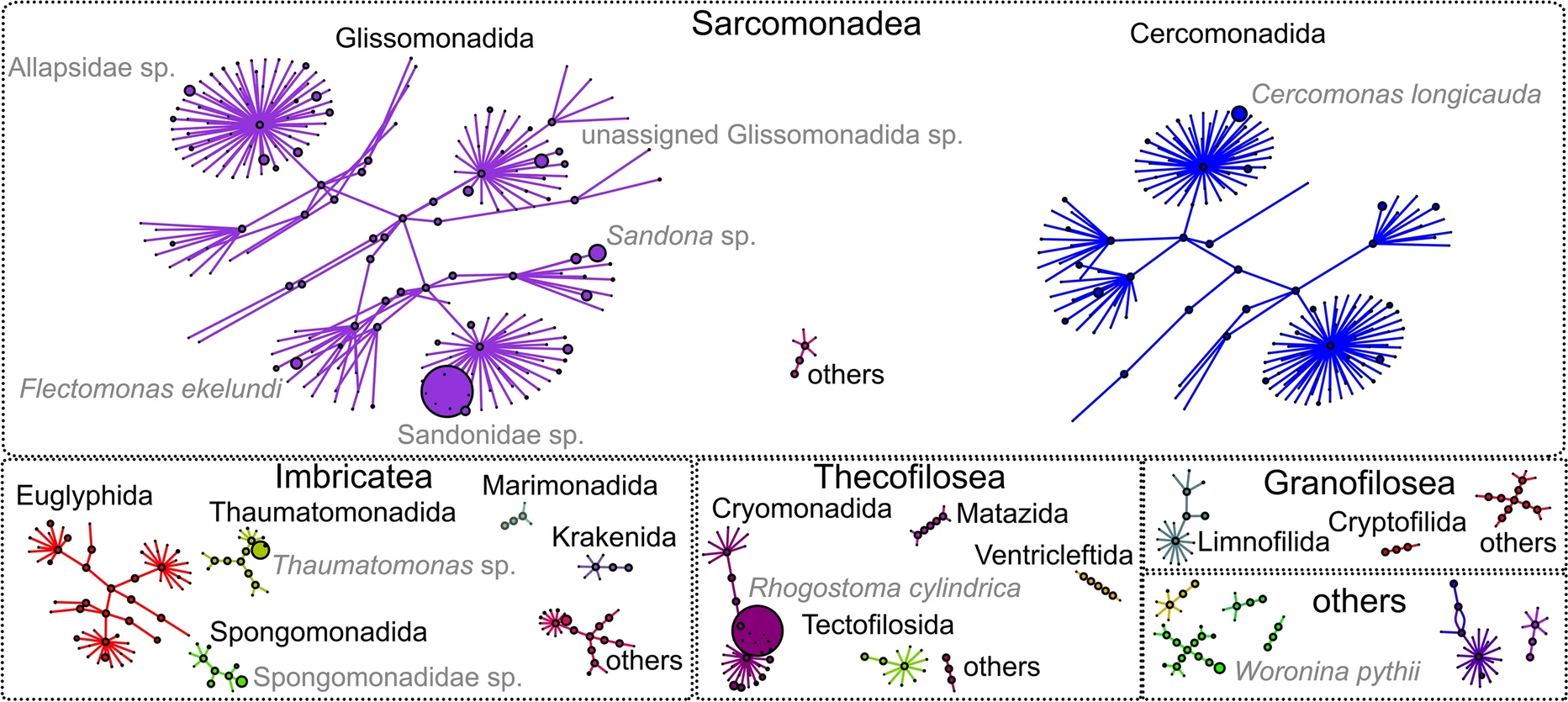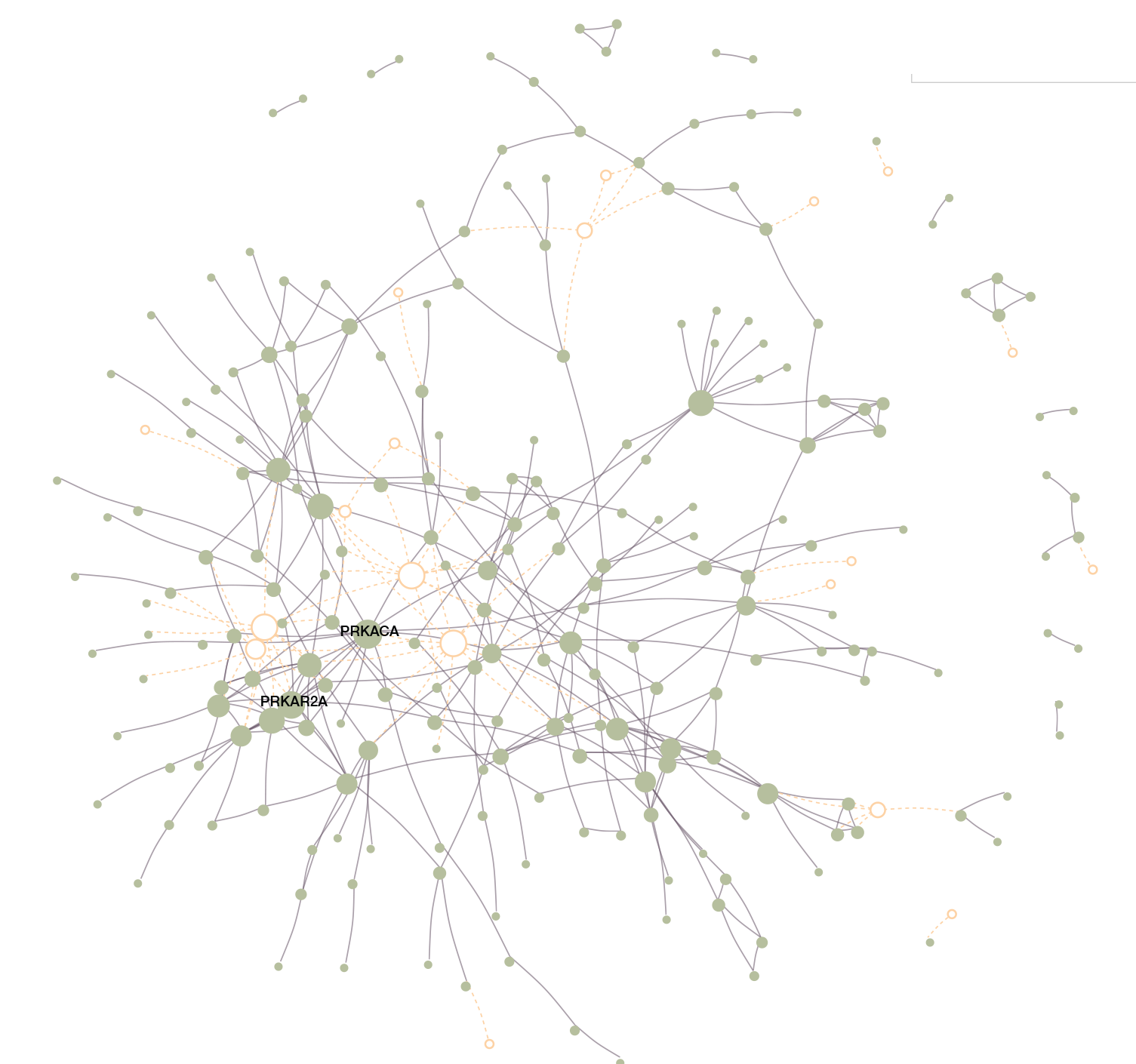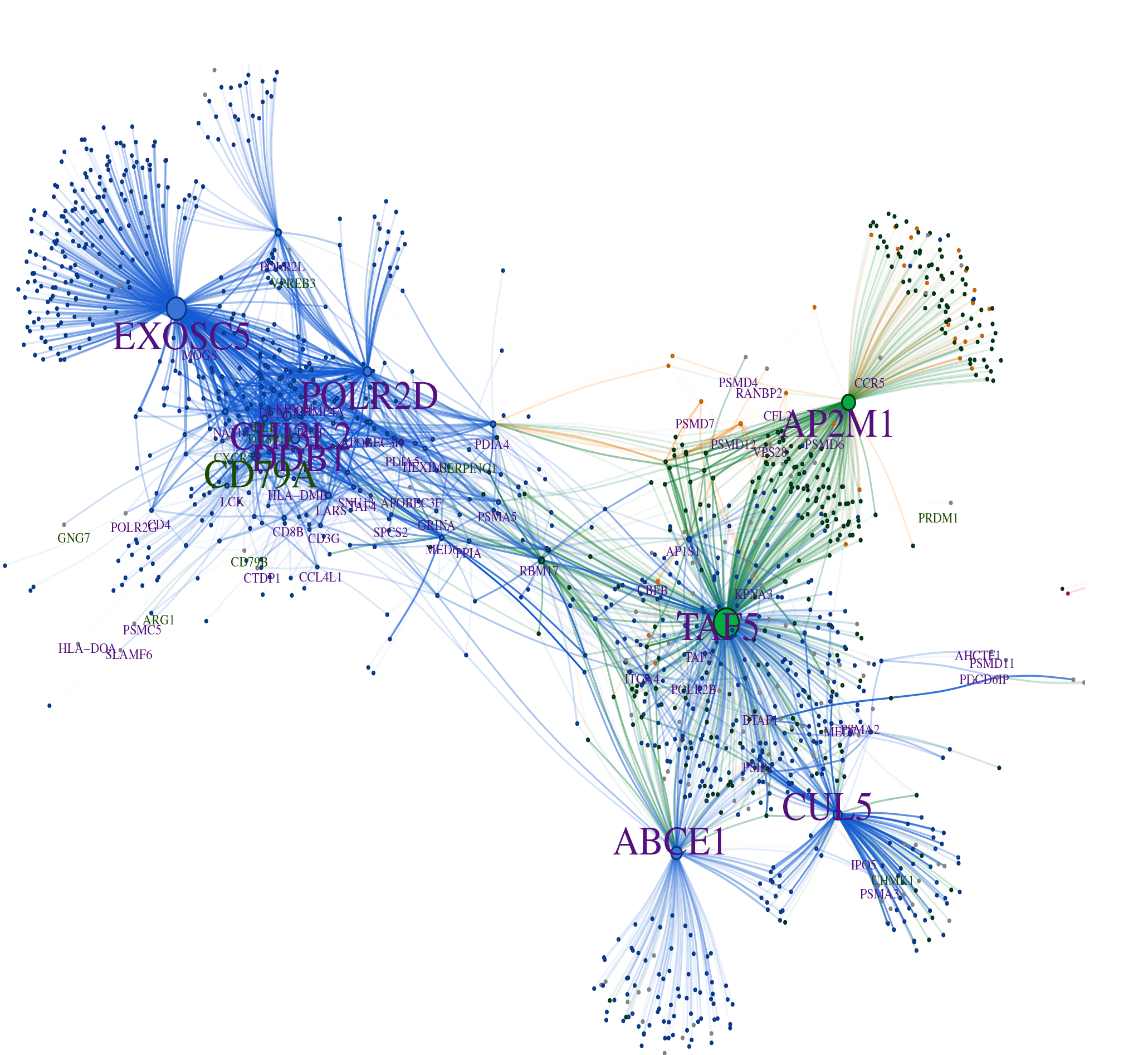
Improving the generalizability of protein-ligand binding predictions with AI-Bind
State-of-the-art machine learning models in drug discovery fail to reliably predict the binding properties of poorly annotated proteins and small molecules. Here, the authors present AI-Bind, a machine learning pipeline to improve generalizability and interpretability of binding predictions.